Research Interests
As a Research Engineer, my main task is to help Researchers in their software developments.
I have the privilege of being involved in several projects spanning a wide range of different questions in the field of Bioinformatics and Computational Biology. Below is a list of the main 4 projects I have been involved in in the last few years.
- Aevol: a digital genetics model (in silico experimental evolution)
- FluoBacTracker: identification and tracking of cells in microscope «movies»
- KisSplice: detection of polymorphisms in RNA-seq data
- SiMuScale: a versatile multi-scale simulator of cell population dynamics
Aevol: a digital genetics model (in silico experimental evolution).
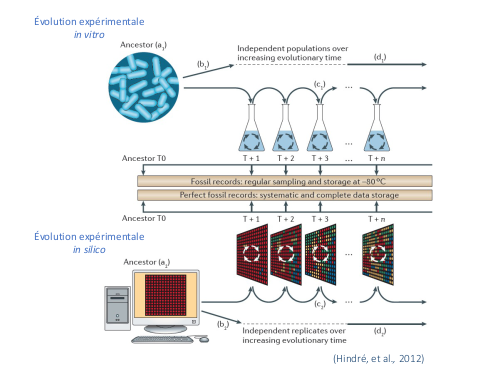
Aevol is a simulation platform for in silico experimental evolution. It was designed to study experimentally the mechanisms responsible for the structuration of the genome and the transcriptome.
Aevol is primarily a digital genetics model: populations of digital organisms undergo a repeated process of variation and selection, which creates a Darwinian dynamic. By modifying the characteristics of variation (mutation types and rates) or selection (population size, spatial structure, environmental changes), one can study experimentally the impact of these parameters on the structure of the evolved organisms. Aevol comes along with a set of tools to analyse both the course of evolution and its outcome.
In Aevol, each artificial organism has its own genome which is evaluated with regard to the environment thanks to a simple artificial chemistry. Because the purpose of the model is to study specifically the evolution of the structure of the genome (e.g. organization of coding vs non-coding sequences, operon structures), the model must be very realistic on that particular level. Not only must the genome itself have a biologically plausible structure, but also the genetic operators: local mutations as well as chromosomal rearrangements and horizontal transfer are hence modelled. The realism of the genome and of the genetic operators is the main specificity of Aevol amongst its competitors.
Website: www.aevol.fr
FluoBacTracker: identification and tracking of cells in microscope «movies».
FluoBacTracker is a image analysis tool that allows for the segmentation and tracking of bacterial cells in a time-lapse microscopy movie. It is easy to set up, has a user-friendly graphical interface and uses an efficient implementation of the CellST algorithm.
Classical computational approaches for the segmentation and tracking of bacteria deal with both problems sequentially, first segmenting all the images and only then addressing the tracking problem. However, in many situations, the segmentation step cannot by done reliably and the corresponding tracking attempt is meaningless. By conducting both tasks at once, the CellST algorithm can combine the knowledge gathered by each and thus enhance both processes.
FluoBacTracker is an ImageJ plugin that implements the CellST algorithm in an efficient and user-friendly way. It keeps track of all the «decisions» that were made during computation and will inject any piece of knowledge the user will provide into the algorithm to minimize manual corrections.
Kissplice: detection of polymorphisms in RNA-seq data.

KisSplice is an RNA-seq data analysis tool that allows for the detection of different kind of polymorphisms in the data both with or without a reference genome/transcriptome.
It is an exact local transcriptome assembler that allows one to identify SNPs, indels, alternative splicing events and other kinds of polymorphisms. It can deal with an arbitrary number of biological conditions, and will quantify each variant in each condition. It has been tested on Illumina datasets of up to 1G reads. Its memory consumption is around 5Gb for 100M reads.
KisSplice is not a full-length transcriptome assembler, it will output only the variable regions of the transcripts rather than reconstructing them entirely. These ouputs can in turn be fed to one of the provided post-treatments depending on the availability of a reference genome/transcriptome and wether a differential analysis is needed.
Website: kissplice.prabi.fr
SiMuScale: a versatile multi-scale simulator of cell population dynamics.
SiMuScale is a C++ simulation framework that models both intra- and extra-cellular processes at different time scales. Its decoupled architecture allows for an easy and parsimonious extension of the model with e.g. a new kind of intra-cellular formalism.
SiMuScale is currently under active development, it will be distributed in the form of a Software as a Service (SaaS) and possibly as a standalone application for specific needs.



